Intronic off-target effects with antisense oligos
Undecided on whether to use silencing RNAs (siRNAs) or antisense oligos (ASOs) for your RNA interference experiments? Read on.
ASOs are single-stranded DNA oligonucleotides which downregulate specific RNA by hybridizing with it, forming a heteroduplex recognizable by RNase H1. RNase H1 is found in the nucleus, hence ASOs are often used to target nuclear-localized RNAs such as non-coding RNA.
A recent paper however highlighted that since RNase H1 is found in the nucleus, intronic sequences may also fall prey to ASO-mediated silencing, exposing ASOs to a much larger array of off-target sequences. Indeed, a quantitative PCR analysis of chemically modified ASOs (marketed as GapmeRs) targeting a gene called BACH1 showed a surprisingly high number of positive off-target effects even with transcripts of low sequence identity (PHF6 – 4 nt gap with exon; 3 mismatches with intron).
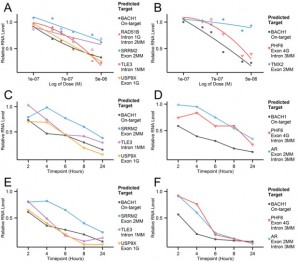
The dose and time-dependent nature of ASO-mediated RNA downregulation (measured by qPCR) in off-target genes of two ASOs targeting BACH1 is seen here in various cell lines.
ASO-mediated off-target effects were detected against both exonic and intronic regions, though intronic regions proved more susceptible. This is illustrated below where greater off-target activity was seen in chart C (intron-based OTEs) as opposed to chart B (exon-based OTEs).
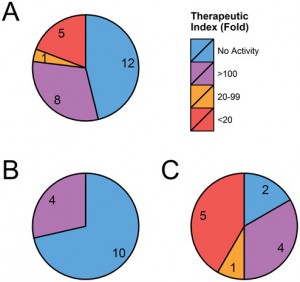
Graphical summary of 26 OTEs predicted and tested for GSK2910546A (ASO). Total (A), exonic (B) and intronic (C) off-target effects are grouped by their potency relative to EC50 of intended target (BACH1, 0.3 μM).
Nuclear-localized RNAs are thought to be less susceptible to siRNA-mediated degradation due to a lower expression level of RNA-induced silencing machinery (RISC) components in the nucleus. That is not to say they are not there. Silencing of nuclear-localized RNAs such as 7SK by siRNA also occurs efficiently, indicating RISC-mediated RNAi can take place in the nucleus.
siPOOLs address hybridization-dependent toxicities and have been shown to work with nuclear-localized RNAs. Contact us to try out siPOOLs today.
Want to receive regular blog updates? Sign up for our siTOOLs Newsletter:
References:
Kamola, P. J., Kitson, J. D. A., Turner, G., Maratou, K., Eriksson, S., Panjwani, A., … Parry, J. D. (n.d.). In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Research , 43 (18 ), 8638–8650. Retrieved from https://nar.oxfordjournals.org/content/43/18/8638.abstract
Gagnon, K. T., Li, L., Chu, Y., Janowski, B. A., & Corey, D. R. (2016). RNAi Factors Are Present and Active in Human Cell Nuclei. Cell Reports, 6(1), 211–221. doi:10.1016/j.celrep.2013.12.013
Robb, G. B., Brown, K. M., Khurana, J., & Rana, T. M. (2005). Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol, 12(2), 133–137. Retrieved from https://dx.doi.org/10.1038/nsmb886





One thought on “Intronic off-target effects with antisense oligos”
Comments are closed.