What is the probability of an siRNA off-target phenotype?
Summary: Conventional siRNAs have a high probability of giving off-target phenotypes. siRNA off-target effects can be reduced by using more specific reagents or narrowing the assay focus (to reduce the number of relevant genes). Even when the assay is relatively focused, more specific reagents significantly increase the probability of observing on-target effects.
Probability of siRNA off-target phenotype depends on reagent specificity and assay biology
The probability of getting an off-target effect from an siRNA depends on several factors, the main ones being reagent specificity and assay biology. If an siRNA down-regulates a large number of genes, or if an assay phenotype can be induced by a large number of genes, the probability of observing an off-target phenotype increases.
siRNAs can down-regulate many off-target genes
Garcia et al. (2011) compiled 164 different microarray experiments measuring gene expression following transfection with siRNAs. The mean number of down-regulated genes in these experiments was 132 and the median was 68 (down-regulated genes were silenced by 50% or more).
As noted in earlier studies of gene expression following siRNA treatment (e.g. Jackson et al. 2003), few of the down-regulated genes are shared between siRNAs with the same target gene. This suggests that the down-regulated genes are not the downstream result of target gene knockdown (i.e. they are mostly off-target).
High-complexity pooling of siRNAs (e.g. with siPOOLs) can reduce the number of down-regulated genes.
The following figure, based on data from Hannus et al. 2014, shows the difference between the gene expression changes caused by a single siRNA (left) and a high-complexity siRNA pool (siPOOL, right), which also includes that same single siRNA:
Estimating the probability of siRNA off-target phenotypes
Assuming different numbers of down-regulated genes (off-target) and different numbers of potent genes involved in assay pathways, we can try to estimate the probability of an siRNA giving an off-target effect.
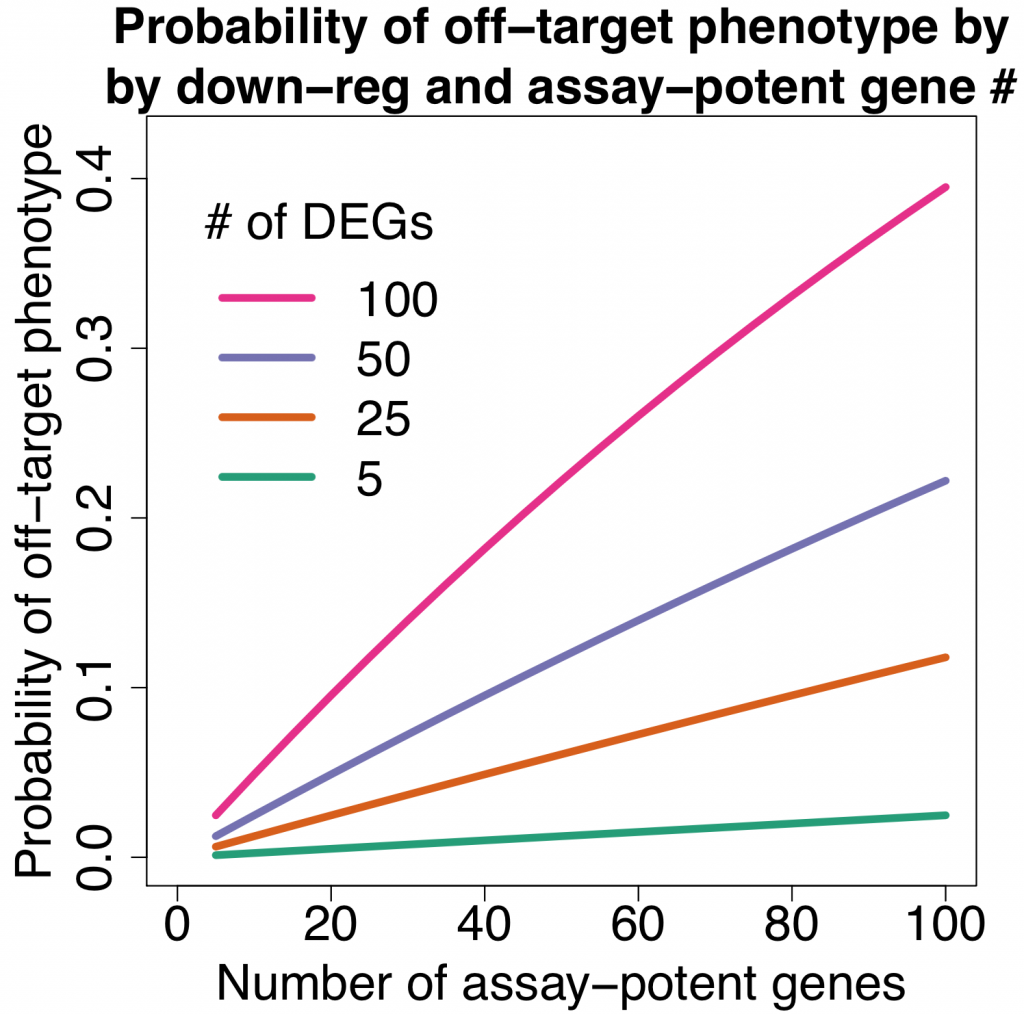
The following plot shows the probability of getting an off-target effect when:
- assuming RNAi reagents down-regulate varying numbers of off-target genes (5, 25, 50, 100)
- down-regulated means that gene expression is reduced by 50% or more
- in the Garcia paper dataset, the mean is 132 and median is 68
- assuming different numbers of assay-potent genes
- an assay-potent gene is one whose down-regulation by 50% or more is sufficient to produce a hit phenotype
- for assays with more general phenotypes (e.g. cell count) we would expect more assay-potent genes
We can see that even if there are only 20 assay-potent genes, there’s a nearly 10% chance of getting an off-target phenotype when siRNAs down-regulate 100 off-target genes (which is close to the average observed in the Garcia dataset).
In a genome-wide screen of 20,000 genes with 3 siRNAs per gene, we would thus expect 2,000 off-target siRNAs.
In contrast, a more specific reagent that only down-regulates 5 off-target genes only has a 0.5% change of producing an off-target phenotype. For the above-mentioned genome-wide RNAi screen, we would expect only 100 off-target siRNAs (a 20-fold reduction).
The importance of RNAi reagent specificity
The above analysis demonstrates the importance of using specific siRNA reagents.
Changing an assay to make the phenotypic readout narrower (to reduce the number of genes capable of inducing a phenotype) is one way to reduce the risk of off-target phenotypes. But this may be a lot of work and is not necessarily desirable or even possible.
A more ideal solution is the use of a specific RNAi reagent, like siPOOLs.
postscript
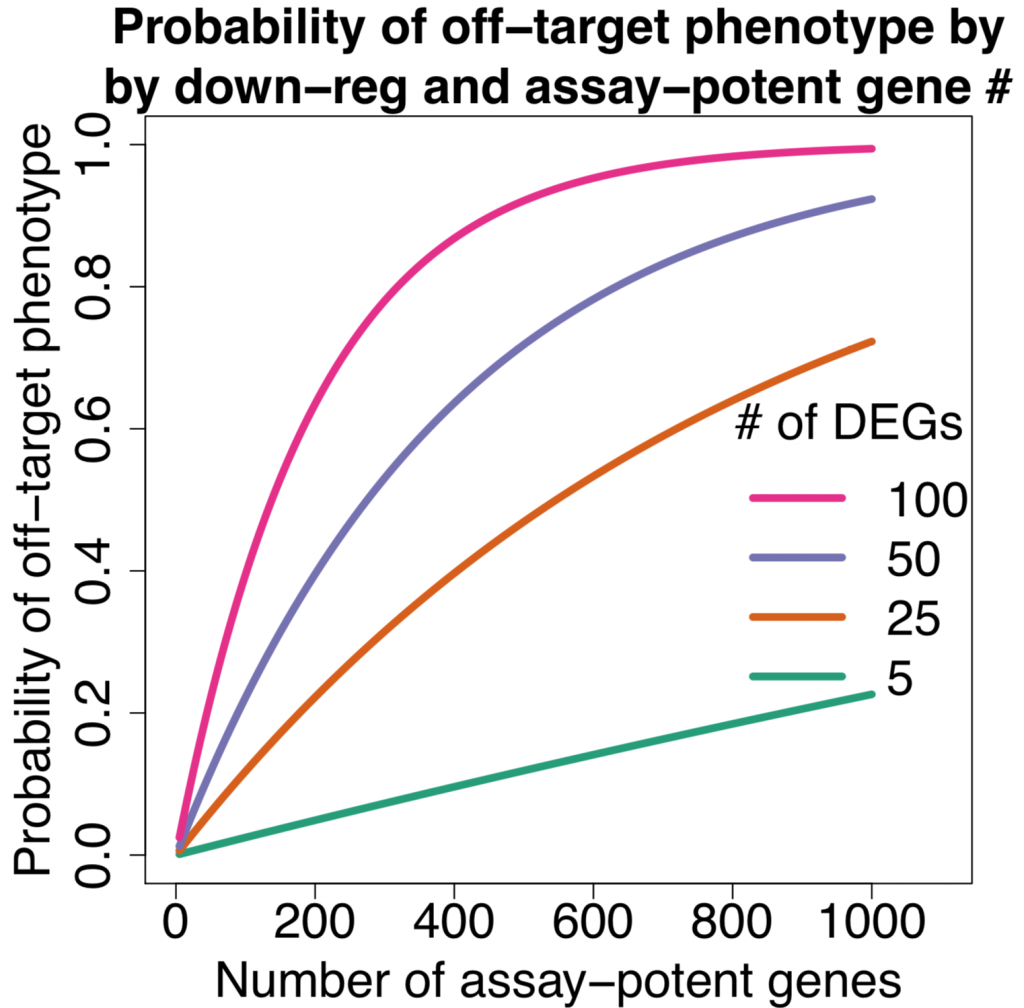
As the number of assay-potent genes increases, the probability of getting an off-target phenotype approaches one.
The following plot (same format as the one above) shows the distribution
The p-values were calculated using the hypergeometric distribution, assuming a population size of 20,000 (the approximate number of protein-coding genes in the human genome).
Note that one of the major simplifying assumptions of the above analysis is that all siRNAs have the same number of down-regulated off-target genes.