Don’t swallow the fly
There was a PI who screened one s i [RNA],
Oh, I don’t know why they screened one s i …
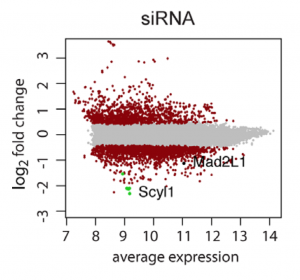
siRNA screens have a high false positive rate, due to pervasive off-target effects.
Confirming ‘hits’ from single-siRNA screens is a lot of work. For low-complexity pool screens, it’s even worse (and, as we will discuss in a later post, less likely to result in true genes of interest).
Progressively, one accumulates a nearly indigestible set of experiments and analyses.
On the in vitro side:
- Screen additional siRNAs for ‘hit’ genes.
- Do quantitative PCR. Single siRNAs vary significantly in their effectiveness, so look for correlation between knock-down and phenotypic strength.
- Create C911 versions of hit siRNAs as off-target controls. To rule out a confounding on-target effect, do qPCR.
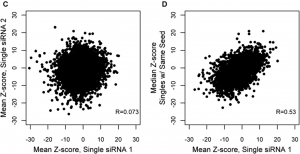
- Screen additional siRNAs with the same seed sequence as off-target controls (as done in a recent paper).
- For low-complexity pools, test the siRNAs individually.
On the in silico side:
- For each hit siRNA, look at plots of phenotypic effects of siRNAs with same seed sequence.
- Adjust phenotypic scores based on predicted off-target effects for seeds.
- Run off-target hit selection tools (like Haystack or GESS), to see if hit genes also show up as strong off-targets.
Does it really have to be so complicated?
Wouldn’t you prefer being able to trust your phenotypic readout?
Better yet, how about hits that don’t turn out to be mostly false-positives?
There is a simpler, better way.
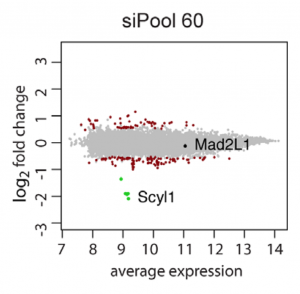
siPools maximize the separation between on-target signal and off-target noise, making interpretation of RNAi phenotypes as clear as possible.