How reproducible are CRISPR screens?
The reproducibility of different CRISPR or RNAi reagents targeting the same gene is sometimes cited as prima facie evidence for the superiority of CRISPR screens to RNAi screens.
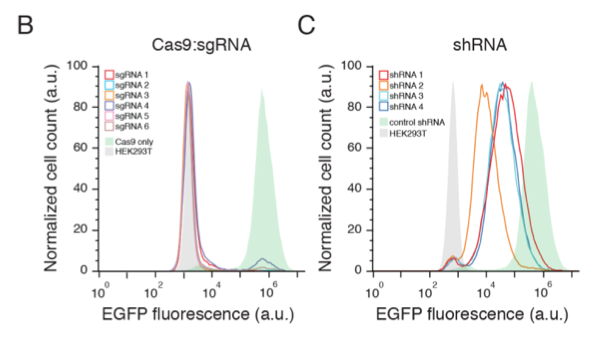
A landmark paper by Shalem et al. showed that different gRNAs inhibit gene expression much more consistently than do different shRNAs:
But does this ensure that CRISPR screens are more reliable (as determined by assay reproducibility) than RNAi screens? Not necessarily.
Shalem et al. performed two pooled CRISPR screens in parallel, and found substantial overlap between the top hits.
How does this overlap compare to that between replicate RNAi screens?
In 2010, Barrows et al. tested the reproducibility between genome-wide siRNA screens conducted 5 months apart. Using the sum of ranks hit selection algorithm, they found 75 and 82 hits from the first and second screens, respectively, with 43 hits overlapping.
If we take the top 75 and top 82 hits from the Shalem replicate screens, we only find 17 genes overlapping.
It’s important to note that the Shalem and Barrows assays were different, as were the screening formats: arrayed (siRNA) vs. pooled (CRISPR). And this was one of the earliest CRISPR libraries. Much has been learned about optimising gRNA efficiency and specificity since the Shalem screen.
However, it is also important to note that consistent inhibition of gene expression does not guarantee consistent phenotypes. The above analysis suggests that care is needed in interpreting the results of CRISPR screens. RNAi screens possess advantages, e.g. ease of arrayed screening, that will make them useful for many years to come.
Want to receive regular blog updates? Sign up for our siTOOLs Newsletter: