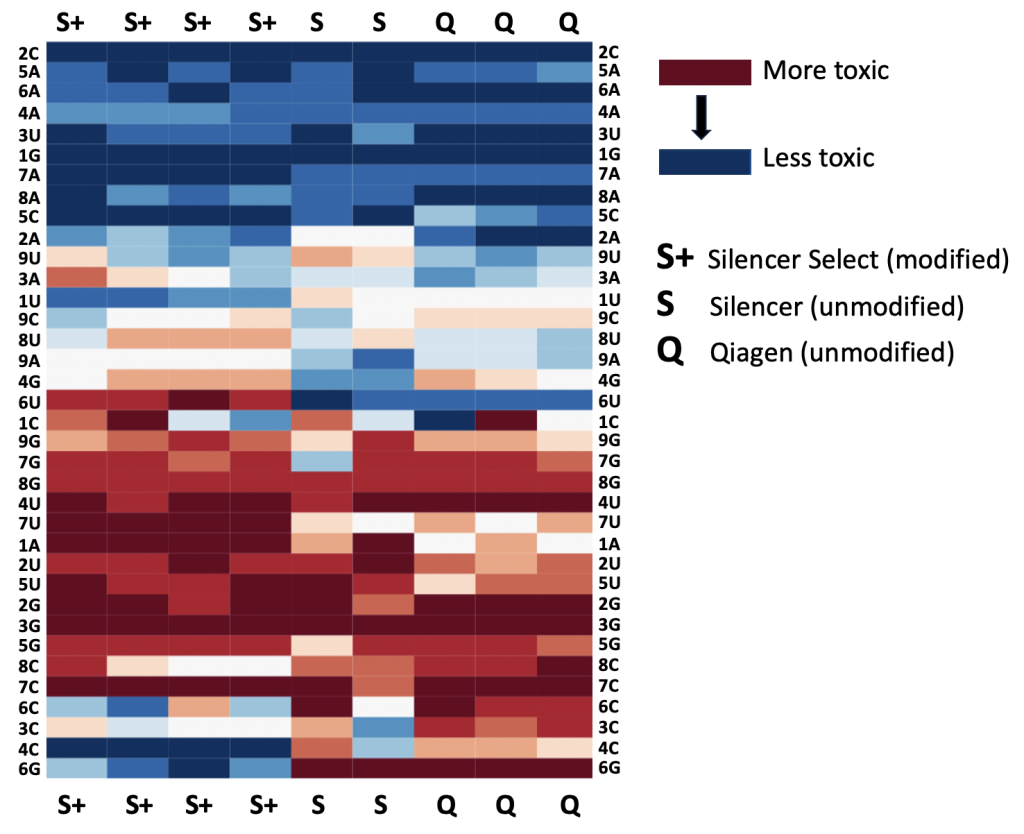
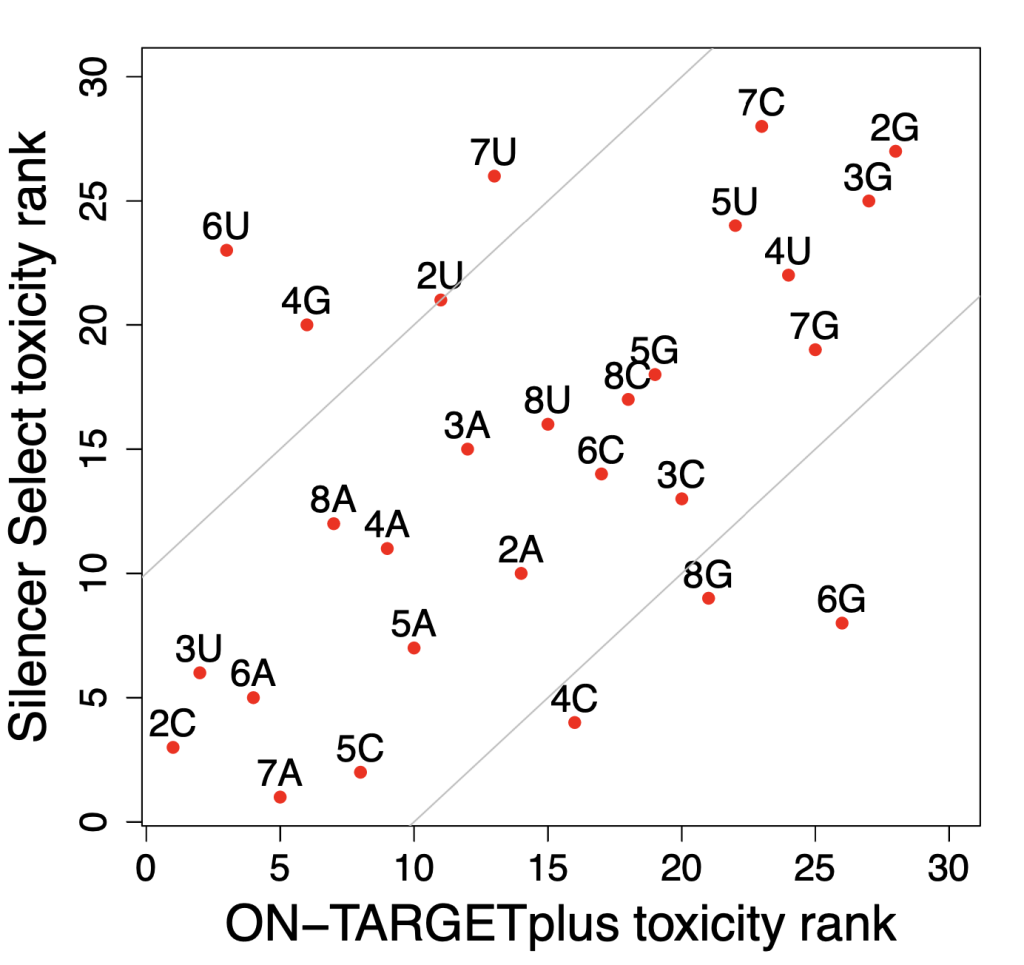
RNAi vs CRISPR: RNAi even better at finding essential genes

Which technology is better, RNAi or CRISPR?
The best answer to this question, like so many others is, it depends.
If cells can adapt and compensate for loss of the gene, or you want to titrate gene levels (important in drug discovery), then RNAi will be better.
If a gene’s transcripts have lots of secondary structure and must be silenced to 99.9% in order to see an assay phenotype, then CRISPR may be better.
We have used two large datasets to attempt to answer the following question: is RNAi or CRISPR better at identifying essential genes?
The first dataset is the BROAD Institute’s Dependency Map (DepMap). It has both RNAi (shRNA) and CRISPR (Cas9) screens from over 700 human cell lines, using hundreds of thousands of reagents. Both types of reagents were used to do pooled screening for cell viability.
The second dataset, also from the BROAD Institute, is called gnomAD. It has genome and exome sequencing for over 100K humans. Based on how frequently mutations are found in the sequenced genomes/exomes (and what type of mutations are preferred), an essentiality score can be assigned to every human gene. It’s the ultimate test (within ethical limits) of whether a gene is essential to humans.
Our approach was as follows:
- for each gene, get the median DepMap viability score across the 700+ cell lines
- done separately for RNAi and CRISPR screens
- for each gene, retrieve the gnomdAD pLI score (probability that loss-of-function not tolerated)
- higher values means the gene is considered more essential
- genes with pLI > 0.9 are classified by gnomAD as essential
If we look at the top 200 genes in each of the RNAi and CRISPR datasets (note: 70 genes are common to both lists), we see that the top 200 genes from RNAi screening are more essential (as measured by pLI) than are the top 200 genes from CRISPR screening. (note that the curves show the running mean for 30 genes)
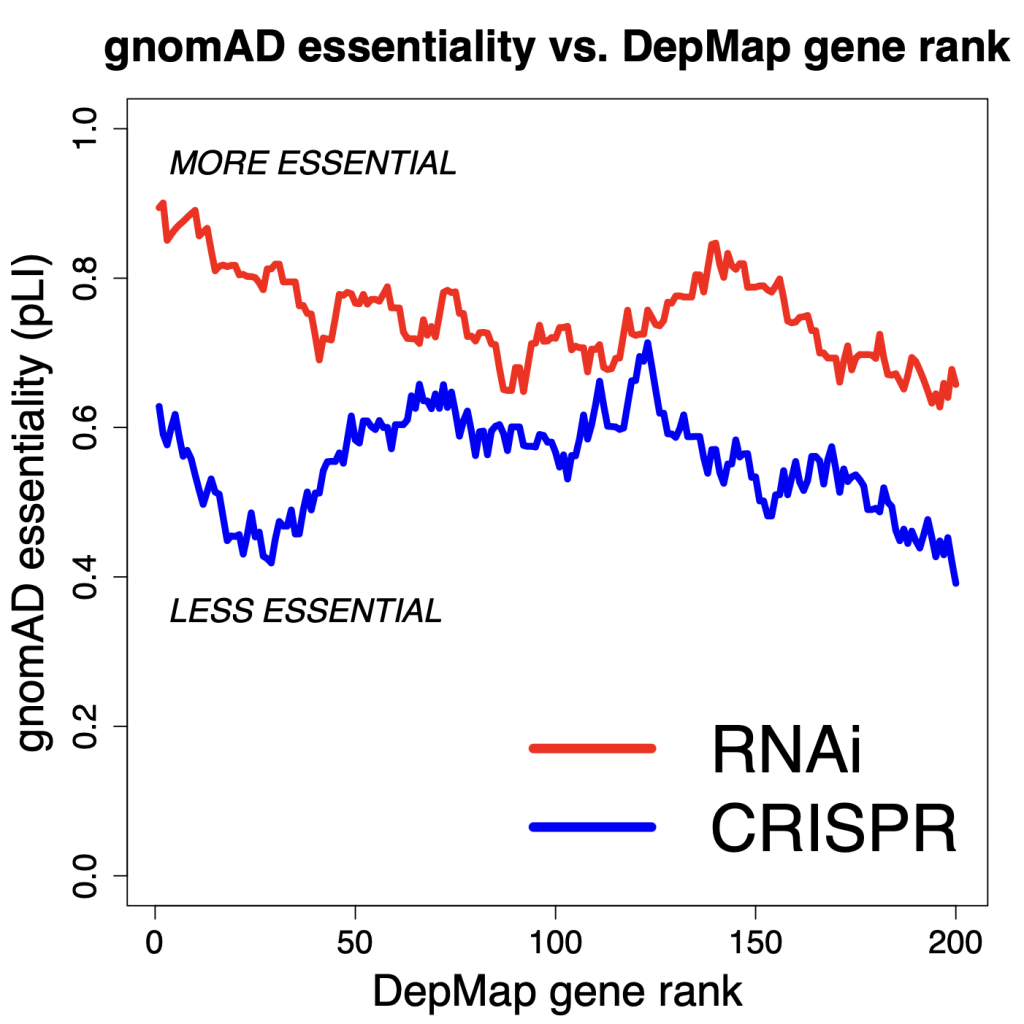
Eventually the curves do converge, but for the top genes, we see that those found by RNAi are more essential.
Alternatively, if we group the genes from the CRISPR and RNAi screens into deciles for cell viability score, we again see that the results from the RNAi screens are more consistent with gnomAD.
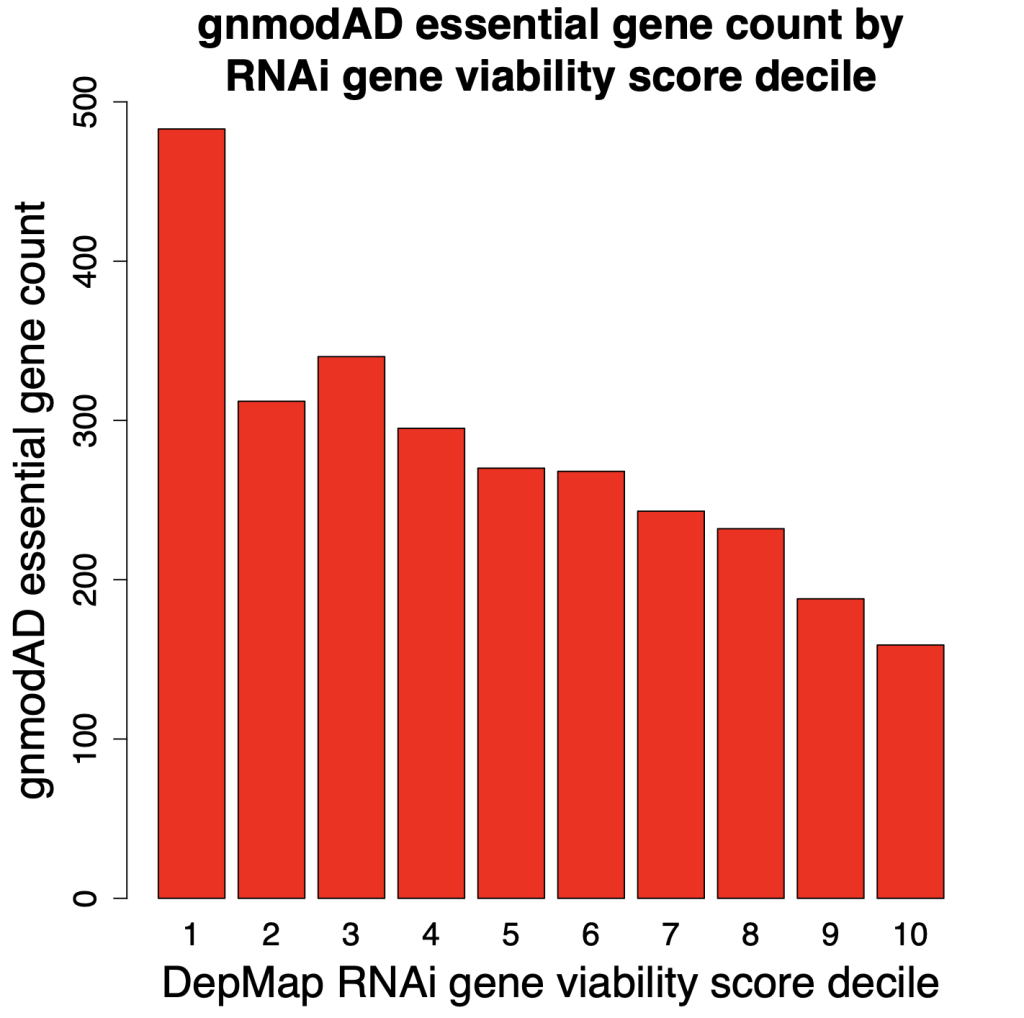
In the following plots, we look at the number of gnomAD essential genes (defined as pLI > 0.9) in each of the deciles. Decile 1 has the top 10% of genes for reducing cell viability (most essential), whereas Decile 10 has the bottom 10% (least essential).
For CRISPR screens, we see that the top 2 deciles show markedly more gnomAD essential genes. But after that, the counts flatten out. There is little difference in the number of gnomAD essential genes in deciles 3 through 10.
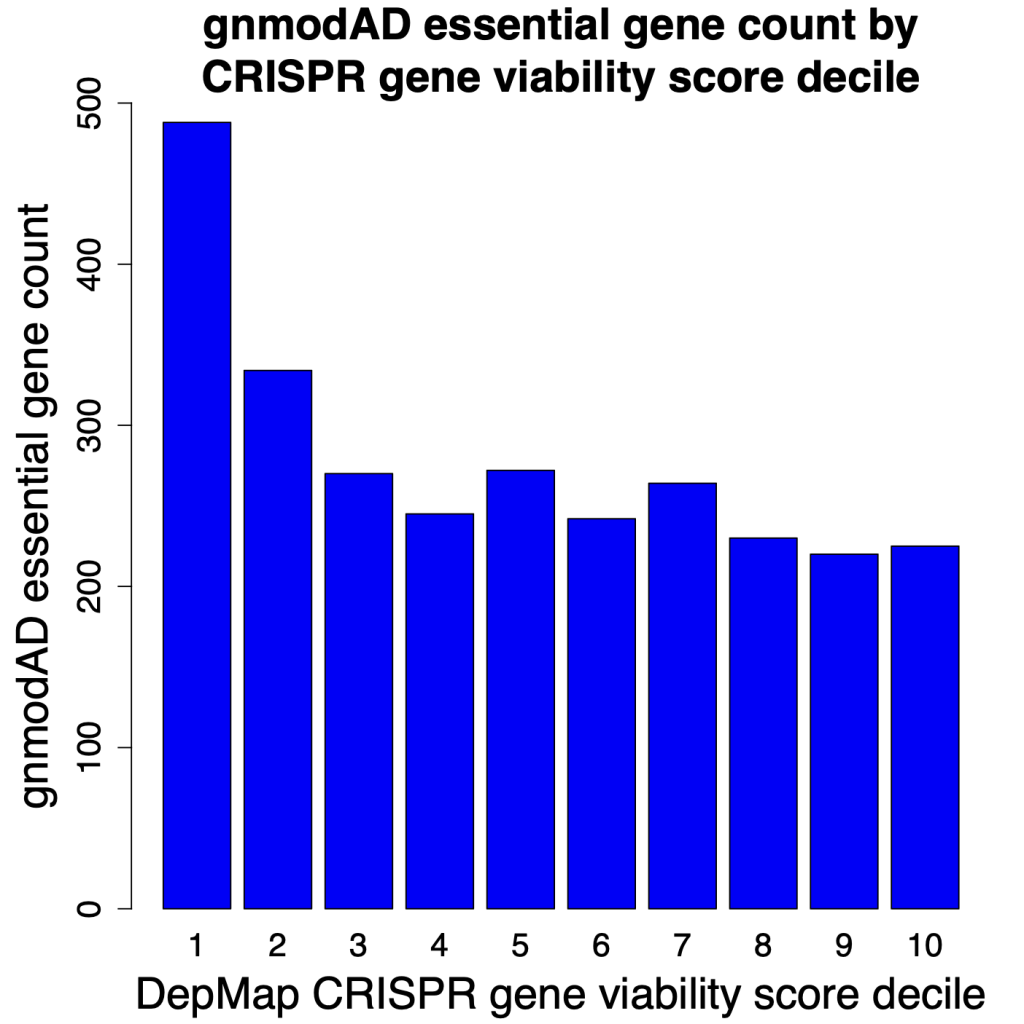
The results from RNAi screening show a fairly steady decline in gnomAD essential genes in deciles 1 through 10. Which is what one would expect. Genes that increase cell count should tend to be less essential. i.e., decile 10 should have the smallest number of essential genes. That is what we see with the RNAi screens, but not with the CRISPR screens.
Conclusion
RNAi and CRISPR screens can both pull out genes found to be essential in the gnomAD dataset.
However, the top genes from RNAi screening tend to be a bit more essential in real-life experiments (i.e., the humans from the gnomAD dataset).
Furthermore, the trend for gnomAD essential gene counts through the ranked datasets makes more sense for RNAi screens than for CRISPR screens.
CRISPR may be a newer technology, but that does not necessarily make it better than RNAi.
Both have their advantages and disadvantages, which we will discuss more in future blog posts.
It should also be noted that two of the main disadvantages of RNAi screening (seed-based off-target effects, and variability in silencing between different siRNAs) have been addressed by siPOOLs.
In an upcoming blog post, we will take a closer look at genes that gave different results in the DepMap RNAi and CRISPR screens.