Complex siRNA pooling is especially important for silencing lncRNAs
One advantage of pooling siRNAs is that the pool tends to silence about as well as any single siRNA. That is why independent complex siRNA pools (siPOOLs) for the same gene have much more similar and better average silencing than single siRNAs or mini-pools (Dharmacon), as discussed in our last blog post.
Another advantage of complex siRNA pools is that you can cover more regions of the target gene. This may be especially important when silencing lncRNAs, which can be very long and whose structure may cause certain regions of the transcript to be inaccessible to RISC.
MALAT1
A good exemplar of this phenomenon is MALAT1, whose transcripts are nearly 9 kb and whose secondary and tertiary structure is important for its cellular function.
Two groups who used single siRNAs or mini-pools (Dharmacon) found poor silencing for MALAT1.
Stojic et al. (2018) used a Lincode mini-pool (Dharmacon) and saw almost no silencing:
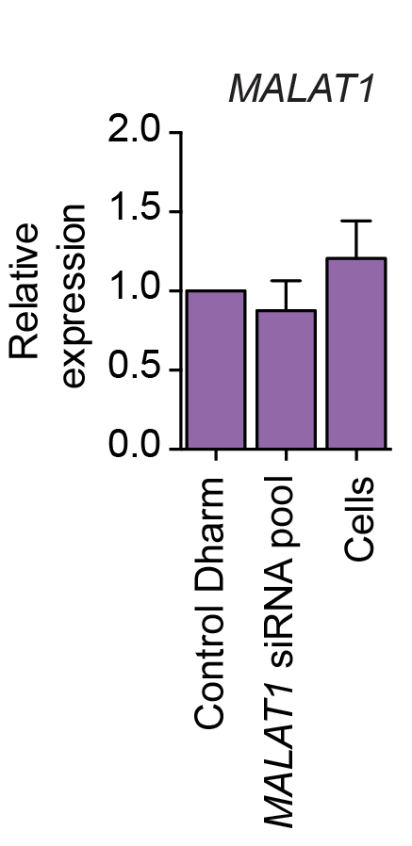
That was despite using a very high siRNA concentration (50 nM) in a cell line where RNAi normally works very well (HeLa).
Lennox and Behlke (2016) used several single siRNAs at two concentrations (1 nM and 10 nM), again in HeLa cells. They found highly variable silencing, where most siRNAs had poor silencing, and a small number worked fairly well at the higher concentration (10 nM):
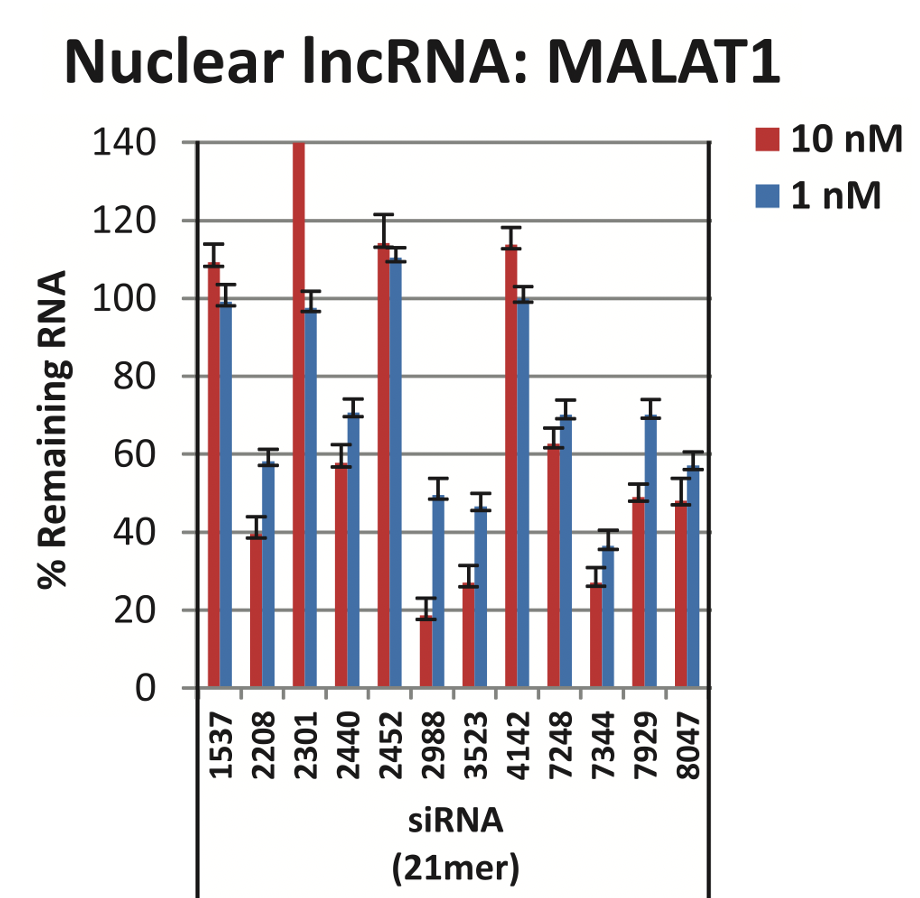
If one were to randomly choose an siRNA, there is only a 25% chance (3 / 12) that it would give decent silencing at 10 nM (using 30% remaining RNA as cutoff for decent silencing). And none would be considered decent at 1 nM.
Of note, this paper is often cited as a reason for not using RNAi for lncRNAs (the authors, from IDT, recommend using ASOs).
The results from Stojic et al. were quite poor (the Lincode mini-pool hardly silenced), and could be due to reagent quality.
siPOOLs effectively silence MALAT1
The results from Lennox and Behlke are more in line with what we’ve observed when researching the silencing of individual siRNAs versus complex siRNA pools (siPOOLs). Some siRNAs are much better than others. And, as mentioned earlier, a complex pool of siRNAs tends to silence like the best constituent siRNAs.
Given the best-silencing-siRNA selection from complex siRNA pooling and the increased transcript coverage, we would expect siPOOLs to give better silencing than what these authors found.
Our 2 independent siPOOLs for MALAT1 give silencing of 16.5% and 24.4% remaining RNA, respectively, at 1 nM:
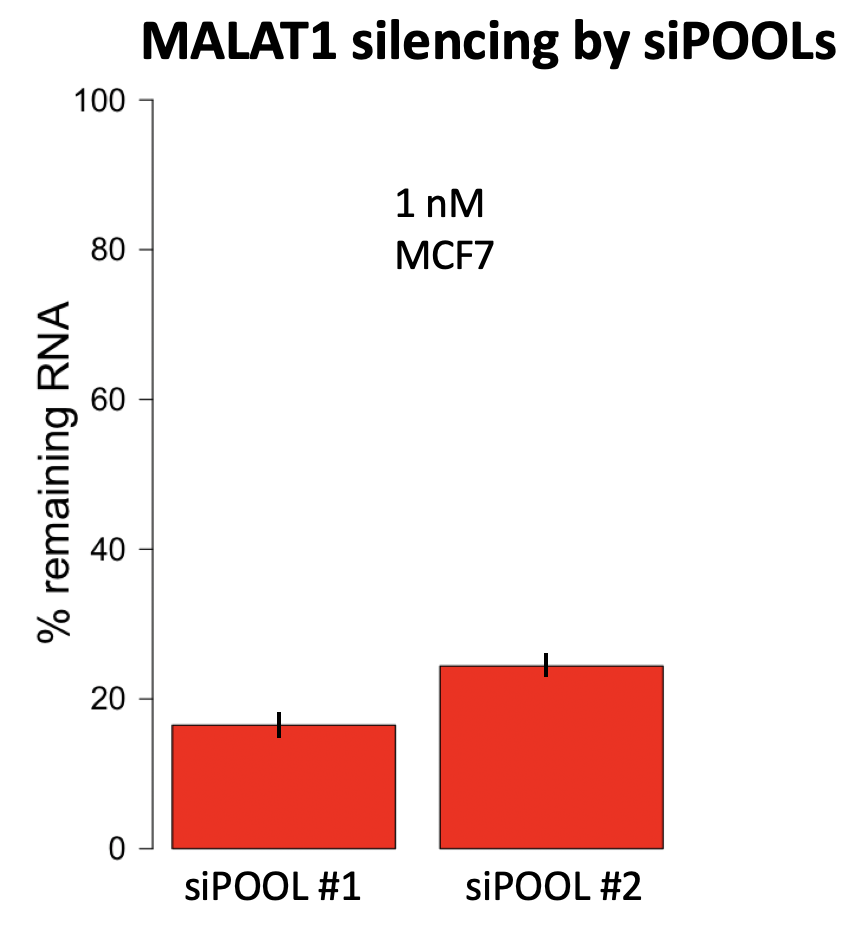
If we compare our reagents to those used by Lennox and Behlke, we see that there is much broader transcript coverage. As mentioned, this could be especially important for lncRNAs like MALAT1 that have extensive secondary and tertiary structure.
Lennox and Behlke siRNAs (used individually):

siPOOL #1 siRNAs (used together):

Conclusion
Two factors make complex siRNA pools (siPOOLs) especially well suited for silencing lncRNAs:
- Complex siRNA pools tend to silence like their best constituent siRNAs. A single siPOOL silences about as well as the best single siRNA. For targets with high silencing variability, siPOOLs are far superior to single siRNAs or mini-pools (Dharmacon).
- By using a complex siRNA pool, more of the gene can be covered. For targets with lots of secondary and tertiary structure, siPOOLs give you the best chance of targeting an accessible region.