ON-TARGETplus siRNAs have strong off-target effects (despite chemical modification)

History of chemical modifications
Chemical modification has long been proposed as a way to limit the off-target effects of siRNAs.
The earliest siRNAs from the two main commercial suppliers (siGENOME from Dharmacon/Horizon Discovery, and Silencer from Ambion/ThermoFisher) were quickly replaced with new chemically-modified siRNAs (ON-TARGETplus from Dharmacon, and Silencer Select from Ambion).
We have already seen that Silencer Select siRNAs, despite their chemical modification, maintain a strong off-target seed effect.
The phenotypic correlation between siGENOME (unmodified) and ON-TARGETplus (chemically modified) low-complexity (4-siRNA) pools for the same gene was shown to be very poor.
However, showing a direct seed effect of ON-TARGETplus siRNAs using published data is not straightforward, since Dharmacon (unlike Ambion) has not made their siRNA sequences publicly available.
Here, for the first time, we show massive seed-based off-target effects from ON-TARGETplus siRNAs.
Seed off-target effects from ON-TARGETplus siRNAs
Tan and Martin (2016) provide a dataset that includes 4 different ON-TARGETplus siRNAs for nearly 700 genes, screened for their effect on nuclear area.
We were also able to find a paper that provides sequences for ON-TARGETplus siRNAs. Those sequences were assigned to the siRNAs from the Tan and Martin screen (details on sequence assignment provided at end of post).
The intraclass correlation (ICC) is a measure of reproducibility of measures of the same group, e.g. siRNAs with the same target gene, or siRNAs with the same 7mer seed.
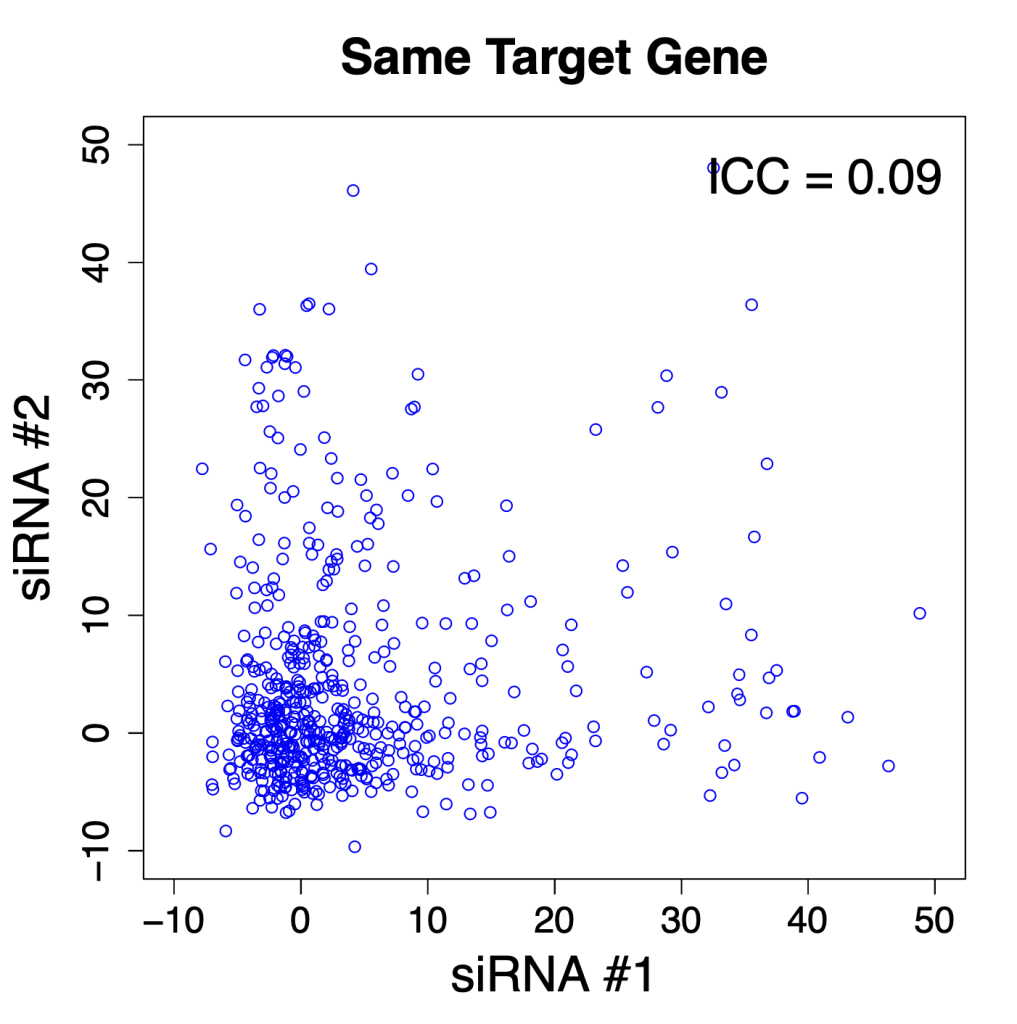
The ICC for ON-TARGETplus siRNAs with the same gene was only 0.09.
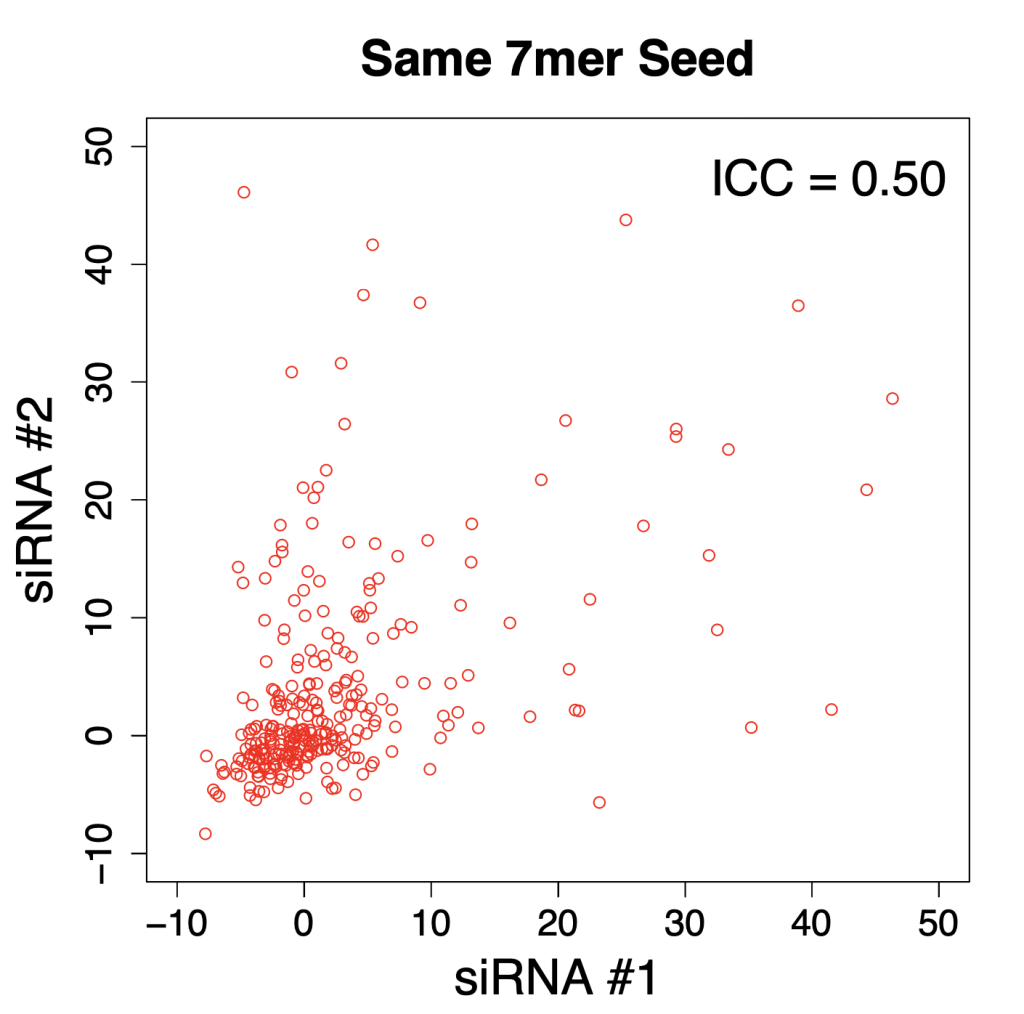
However, the ICC for ON-TARGETplus siRNAs with the same 7mer seed was much higher: 0.50.
Despite chemical-modification, the phenotype of ON-TARGETplus siRNAs is still mostly driven by off-target seed effects.
To show these ICCs graphically, here is a plot with pairs of siRNAs for the same target gene (2 of 4 siRNAs chosen randomly for each gene). [ note that some outliers were removed to assist comparison with same-seed siRNAs]
And here is the plot with pairs of siRNAs with the same 7mer seed:
Conclusion
Chemical modification does not get rid of seed-based off-target effects.
The only effective way to robustly eliminate these effects is with high-complexity (30+ siRNA) pools (siPOOLs).
Technical notes
In order to determine the sequence of the ON-TARGETplus siRNAs from the Tan and Martin screen, the sequences from the supplementary materials of Kim et al. were assigned in order to the siRNAs sorted by catalog number. It is possible that some of the sequences thus assigned were not correct (e.g. Tan and Martin may have used different siRNAs from those listed in Kim et al. for some of the genes), in which case the observed seed effect is actually underestimated.